
What is BioCompute?
Tremendous insights can be found in genome data, and many of these insights are being used to drive personalized medicine. But the hundreds of millions of reads that come from a gene sequencer represent small, nearly random fragments of the genome that’s being sequenced, and there are countless ways in which that data can be transformed to yield insights into cancer, ancestry, microbiome dynamics, metagenomics, and many other areas of interest.
Because there are so many different platforms and so many different scripts and tools to analyze genome data, there is a great need to standardize the way in which these steps are communicated. The more analysis steps and the more complicated a pipeline, the greater the need for a standardized mechanism of communication. The BioCompute standard brings clarity to an analysis, making it clear and reproducible.
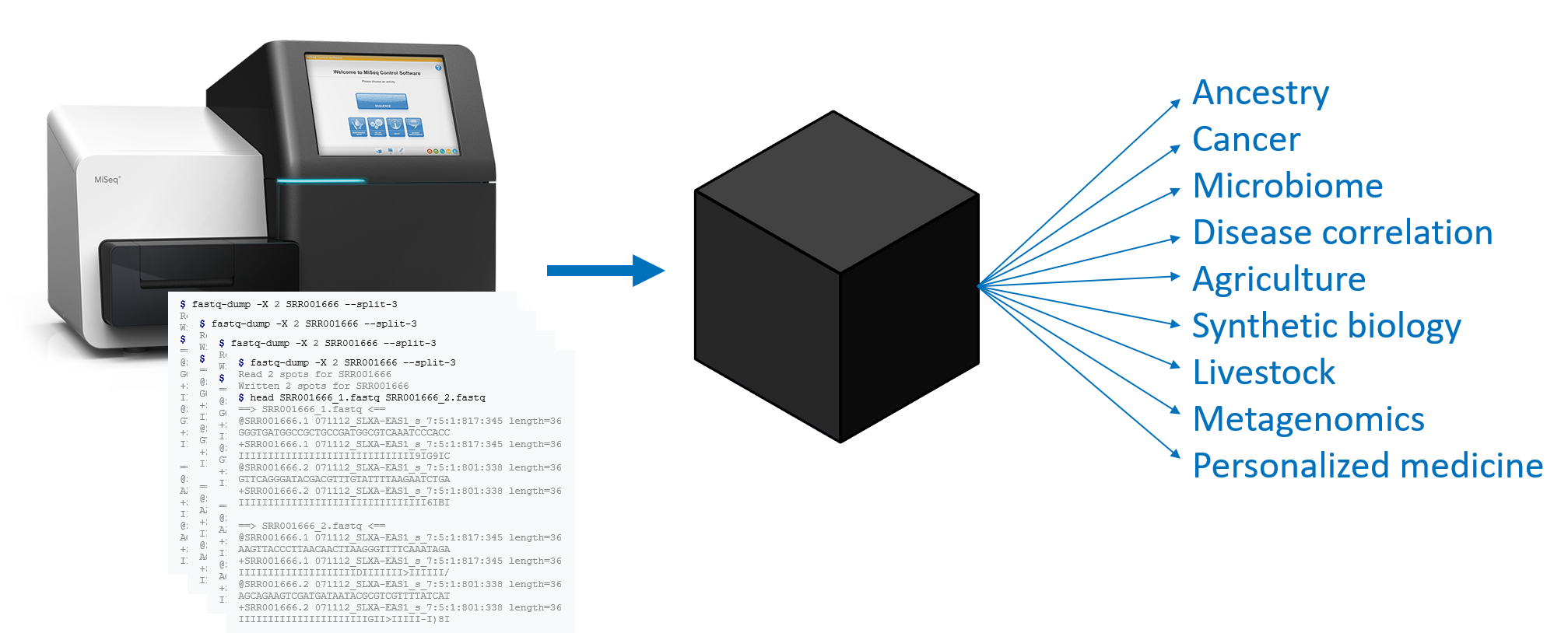
A BioCompute Object (BCO) is an instance of the BioCompute standard, and is a computational record of a bioinformatics pipeline. A BCO is not an analysis, but is a record of which analyses were executed and in exactly which ways. In this way, a BCO acts as an interface for existing standards. A BCO contains all of the necessary information to repeat an entire pipeline from FASTQ to result, and includes additional metadata to identify provenance and usage.
WiFi Analogy
The 802.11 standard (more commonly called “WiFi”) is a way of standardizing communication between vastly different products on a wireless network. If a product manufacturer wants a product to be able to communicate on a wireless internet network, they can configure the device to use the WiFi standard and it will be able to communicate with most commercial routers, regardless of whether the product is a Mac, a PC, a cell phone, or a smart toaster.

BioCompute fills a similar need. BioCompute is not an automation or a new programming language, it is a way of collecting and communicating information between two entities. Rather than a latop and a router, it may be between a pharmaceutical company and the FDA, or between two clinicians, or between a clinician and a researcher. In much the same way that WiFi does not standardize the data that’s being transmitted – allowing you to use Apple’s Facetime, Microsoft’s Internet Explorer, or your favorite cell phone app – BioCompute does not standardize the platforms or tools that are used for genome analysis. You continue to use your favorite platforms and tools, whether it’s HIVE, Galaxy, Seven Bridges, DNAnexus, or others. Also like WiFi, BioCompute can be layered with other privacy or security protocols depending on usage. So clinical trial data can be secured and HIPAA-compliant, while government-funded data sets shared between researchers can be completely open access.
Because BioCompute acts like an envelope for an entire analysis pipeline, it is compatible with other existing standards, including FHIR Genomics and GA4GH.
BioCompute Description
BioCompute is written in Javascript Object Notation (JSON), which is simply a set of key:value pairs (meaning that raw files can be read without any knowledge of programming). Information within the BCO is organized into “domains.” The domains within a BCO record are Provenance, Usability, Extension, Description, Execution, Input/Output, and Parametric Domains. For more information on the domains, please see the BioCompute Schema.
BioCompute was built through a collaboration between The George Washington University and the FDA to improve communication of bioinformatics pipelines, and has since been expanded and refined through the participation or collaboration of hundreds of participants from throughout the public and private sectors. While we welcome interest and membership from anyone, most users will fall into one of three categories:
Research Community
The Biocompute standard can help substantially improve replicability, making it possible to repeat a pipeline on a different sample with high fidelity and high confidence.Clinical Community
As BioCompute Objects become tested and validated, they can be applied in the clinic to identify risk factors, flag pharmakogenetic information, and much more.Pharma, Biotech and Regulatory Pipeline
Protracted communications with the FDA can extend the review process by months. A standardized method of communicating HTS data may help repeat results more quickly and without the need for additional communication.
Research, clinical, and regulatory groups are key drivers of personalized medicine that is based on next generation sequencing, but there are barriers between these groups. BioCompute reduces these hurdles and brings transparency to the workflow, making it more clear what was done, and clearly delineating expectations for data sharing. The BioCompute specification can be layered with other privacy and security protocols to guard sensitive data, or be made open source depending on the needs of the user.
The BioCompute project has generated two publications, three workshops, FDA funding, contributions from over 300 participants, and FDA submissions. The project has worked with individuals from NIH, Harvard, several biotech and pharma companies, EMBL-EBI, Galaxy Project, and many more, and can be integrated with any existing standard for HTS data. The project is expected to be both an IEEE and ISO recognized standard within 8-10 months.
More information about The current BioCompute standard can be found on the Open Science Foundation website (where the standard is developed and maintained), the HIVE website, and the Research Objects discussion of BioCompute.
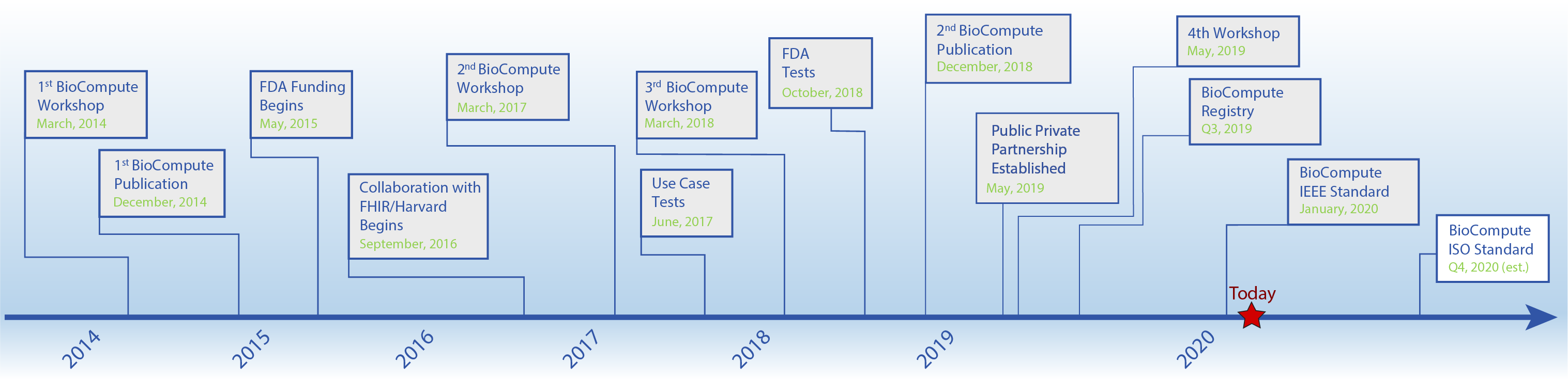
Milestones in the BioCompute Program
The major milestones of the BioCompute Partnership and future goals are paving the way for a consensus-driven, widely adopted standard. The FDA’s Genomics Working Group (GWG) originally articulated the challenges of communicating genomic analysis pipelines in a regulatory context in 2013. Since then, the project has accumulated tremendous momentum, a testament to the GWG’s efforts in describing communication challenges. More recently, the second BioCompute publication has recently been published, the 4th Workshop is scheduled, and the next major goal is the formal launch of the BioCompute Public Private Partnership. The Executive Committee will formalize the future roadmap beyond these goals.
GWU Members

Raja Mazumder
Co-Founder
The George Washington University, DC

Jonathon Keeney
Project Lead
The George Washington University, DC

Charles Hadley King
Technical Lead
The George Washington University, DC

Tianyi Wang
Outreach Lead
The George Washington University, DC
External Collaborator

Dennis Dean
Seven Bridges Genomics, MA

Jeremy Goecks
Georgia Institute of Technology, GA

Carole Goble
University of Manchester, UK

Gil Alterovitz
US Despt of Veterans Affairs, DC

Vahan Simonyan
CRISPR Therapeutics, DC

Jonas Almeida
National Cancer Institute, MD

Dan Taylor
Internet2
Anton Golikov
Food and Drug Administration, MD

Ntino Krampis
The City University of New York, NYC

Michael Crusoe
Common Workflow Language

Stian Soiland-Reyes
University of Manchester, UK

Konstantinos Karagiannis
Food and Drug Administration, MD

Elaine Thompson
Henry M Jackson Foundation for the Advancement of Military Medicine, MD

Nicola Soranzo
Galaxy Platform, UK
Former GWU Members

Janisha Patel
George Washington University, DC
Former Collaborators

Ogan Abaan
Illumina

Payal Banerjee
National Human Genome Research Institute

Amanda Bell
Frederick National Laboratory for Cancer Research

Surajit Bhatta-charya
Children's National Hospital

Lee Black
Tempus Labs, Inc

Ben Busby
DNAnexus

Kristy Cloyd-Warwick
DNAnexus

Ryan Connor
National Center for Biotechnology Information

Paul Duncan
The Janssen Pharmaceutical Companies of Johnson & Johnson

Josep Gelpi
University of Barcelona

Jeremy Goecks
Oregon Health and Science University

Jonathan Jacobs
ATCC

Xiandong Meng
University of Science and Technology

David Michaels
George Washington University

Hiroki Morizono
Children's National Medical Center

Rahi Navelkar
Harcard Medical School

Asa Oudes
Benchling

Jason Travis
TGen

Paul Walsh
Accenture

Jianchao Yao
Merck

John Penn
DNAnexus

Megan PotterSbusch
-

Jonathan Pryke
AstraZeneca

Dan Taylor
Internet2