This document is part of the BioCompute Object User Guide
Creating a BCO on Galaxy
Galaxy has an extensive workflow system. They define a workflow as “.. a series of tools and dataset actions that run in sequence as a batch operation”. An instance of a Galaxy workflow is known as an “invocation”. Since a BCO is a record of a specific instance of computation, the Galaxy invocations are used to generate the Galaxy BCOs.
The first step in creating a well-documented Galaxy BCO is to follow the Best Practices for Maintaining Galaxy Workflows.
Galaxy workflow attributes used by BCO
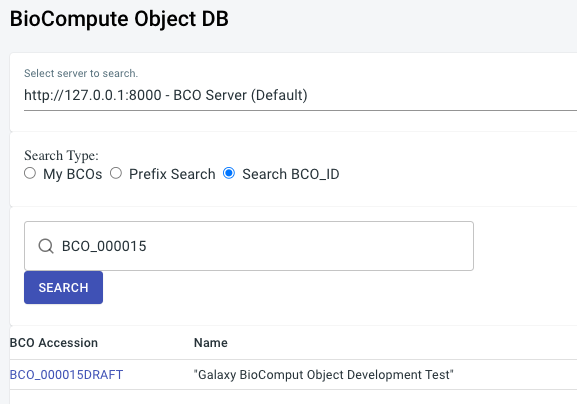
Proper annotation of the workflow is required BEFORE running it. Notice the following in the example below:
- The Name of the workflow becomes the BCO Name
- The workflow version translates to a sequential digit and is included as the BCO version
- The workflow Annotation block becomes the FIRST entry in the BCO Usability Domain
- The annotation from the history (if included) becomes the second entry in the BCO Usability Domain
- The workflow License becomes the BCO License
- The workflow ‘creator/s’ are added to the BCO contributors specification
- The workflow Tags are added to the BCO description domain tags
- It is also possible to annotate each workflow step or tool individually. Depending on the tool or step this information will also be included.
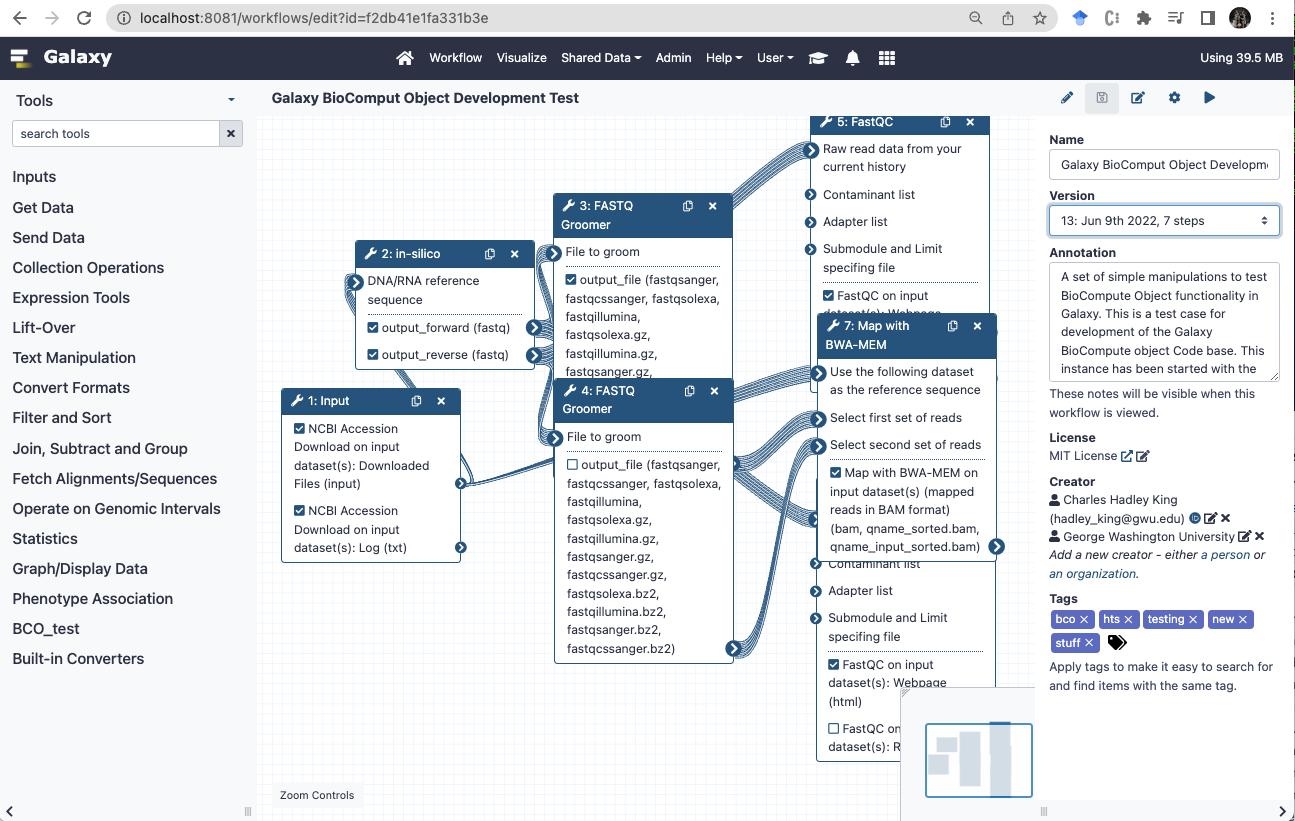
Galaxy history attributes used by BCO
- The Galaxy history will also contribute annotation and tags to the BCO
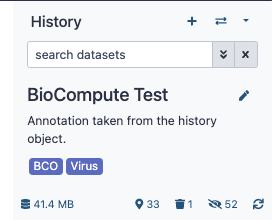
BCO Export from Galaxy
There are two methods for exporting a BCO from Galaxy. Download and Submission to a BCODB via API.
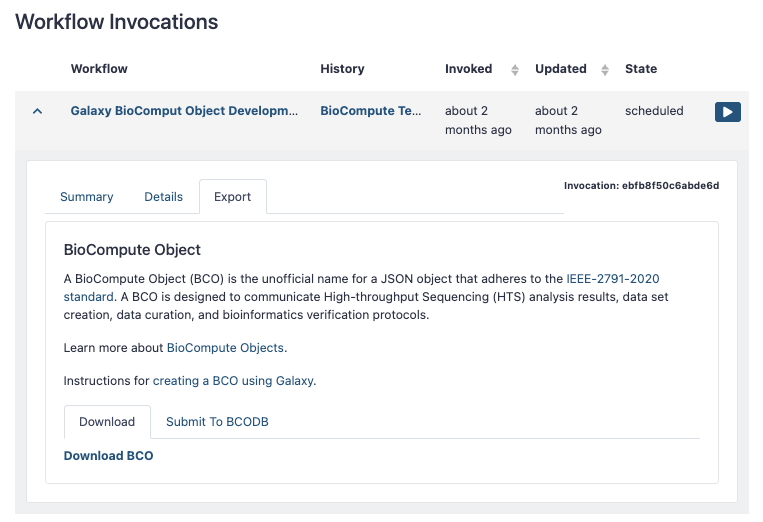
- From the User menu select “Workflow Invocations”
- Expand the invocation you would like to convert to a BCO
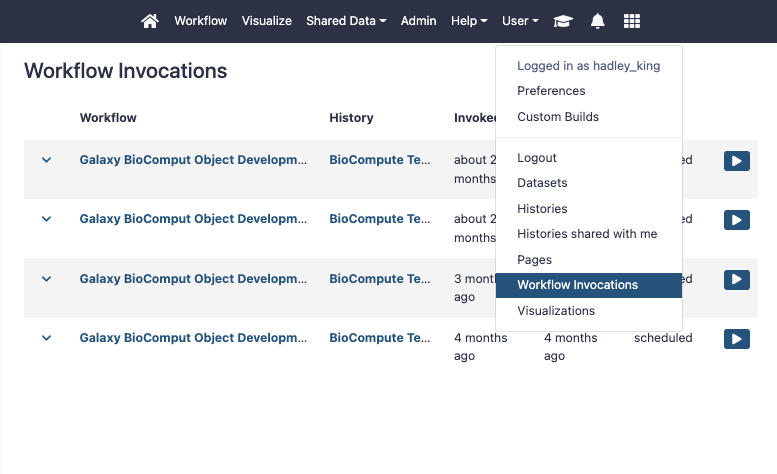
- Select the “Export” tab
Download BCO
Downloading a BCO from Galaxy will save the raw JSON file to your local drive.
- Click “Download”
Submission to a BCODB via API
Submission to the BCODB requires that a user already has an authenticated account at the URL they wish to submit to. Once an account is verified you will need four pieces of information to submit the BCO.
BCODB Root URL
This is the URL for the BCODD that you are submitting your object. In most cases, it will be the public server at ‘https://biocomputeobject.org’.
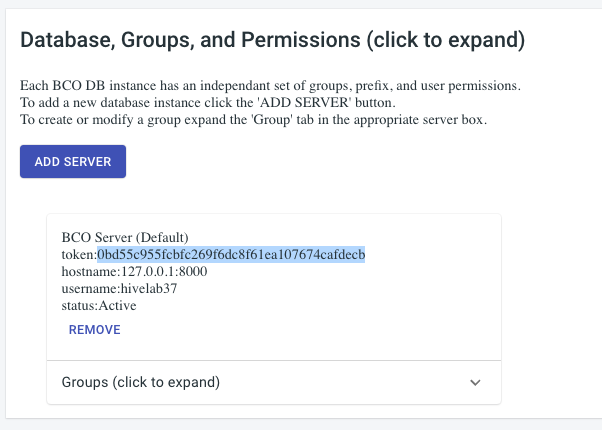
BCODB User API key
The BCODB User API key OR user Token is found on the account page. Expand the “Database, Groups, and Permissions” tab. You will find the token listed in the card for the server you want to submit to.
BCODB Prefix
The BCO Prefix is explained here in the section ‘2.0.2 BioCompute Object Identifier “object_id”’. The default prefix “BCO” is for general use. Other Prefixes require special permission. Please see the BCO Prefix Registry for more information.
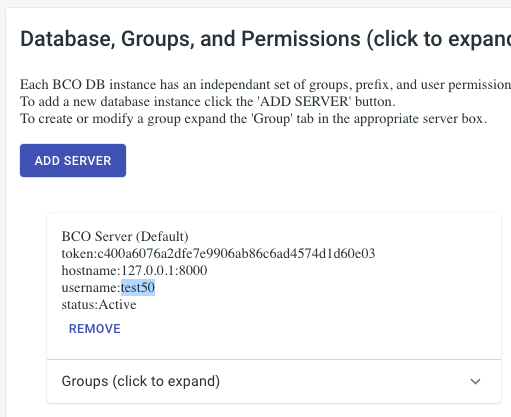
BCODB User Name
The BCODB User Name is found on the account page. Expand the “Database, Groups, and Permissions” tab. You will find the username listed on the card for the server you want to submit to, just like the token.
Submission and next steps
The completed form should look something like the one below:
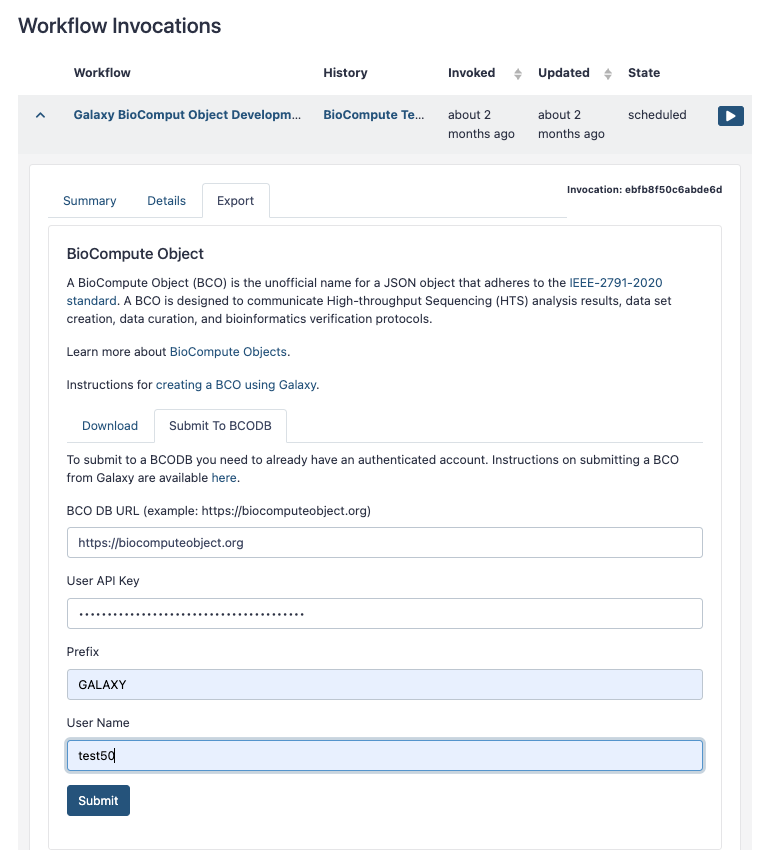
Upon successful submission you should see the objct_id for the BCO just created:
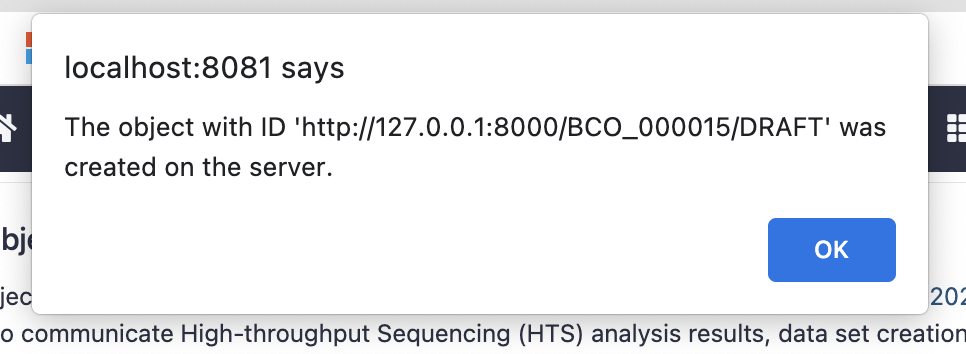
To find the object and modify further or publish go to the BCO Portal and search for the identifier Galaxy gave you.